Tutorial 3. Test phylogenetic signal
Summary:
Phylogenetic signal is the pattern we observe when close relatives are more similar than distant relatives with respect to a particular trait. This tutorial describes the use of RASP to examine primate characters and sociality in a phylogenetic perspective as Kamilar and Cooper (2013). It will take you through the process of states, estimating phylogenetic signal in each trait, and ancestral state reconstruction for sociality of primates.
Information of data source
The consensus tree, 100 trees dataset and 7 genes were downloaded from the 10k Trees Website (https://10ktrees.nunn-lab.org, Arnold et al., 2010). The sociality character is modified from Shultz et al. (2011), in which A=solitary, B=pair-living, C=uni-male and D=muti-male. Other characters are modified from Kamilar and Cooper (2013).
Please notice that we use less than 1/5 of the original dataset as an example, therefore the result may not be exactly the same as the original paper.
ALL files used in this tutorial are stored in examples/Primate/. If you are a beginner of RASP, please start from Tutorial 1.
Loading the data files
Click [File> Close Current Data] or reopen RASP to clear the current trees.
Open [File > Load Trees> Load Trees (more format)] and navigate to Trees_States/gene_trees and select all seven tree files.
Open [File> Load consensus Tree> Load User-specified Tree] and navigate to Trees_States /Primates.tree and select it.
Open [File > Load States (Distributions)], navigate to States/characters.csv and select it.
You will see a window like this:
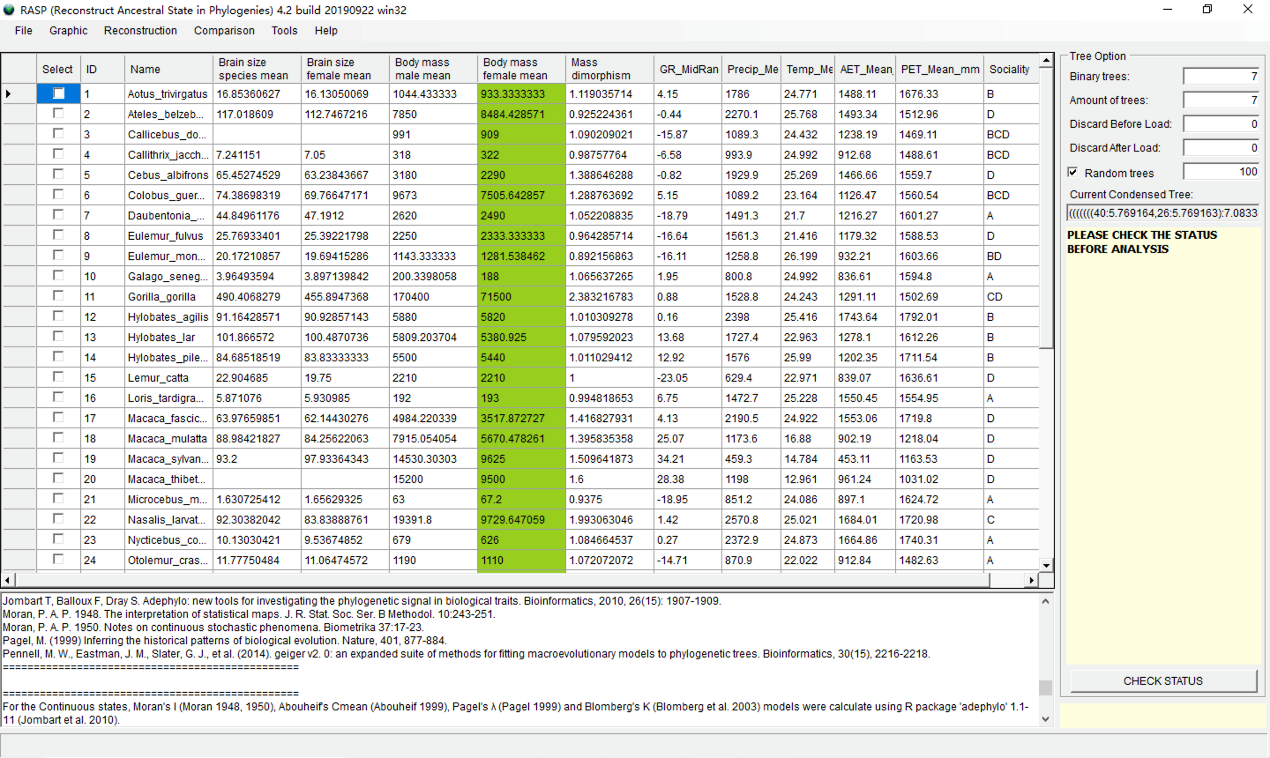
Click the head of column to select it. In this part, we selected the "Body mass female mean" as an example.
NOTE: When some values of characters are missing, please just leave them blank.
Estimate phylogenetic signal on selected state with different genes
In RASP, we use the R package 'adephylo' (Jombart et al. 2010) and 'geiger' 2.0.6.2 (Pennell, et al. 2014) to estimate phylogenetic signal.
Open [Comparison> Trees & States], you will see a window like this:
Click [Analysis> State vs. Trees] to begin the analysis. You may see a command window (in Windows) with some additional information. Keep it open until it closes automatically. This analysis took 30 seconds to run. When finished, you will see a window like this:
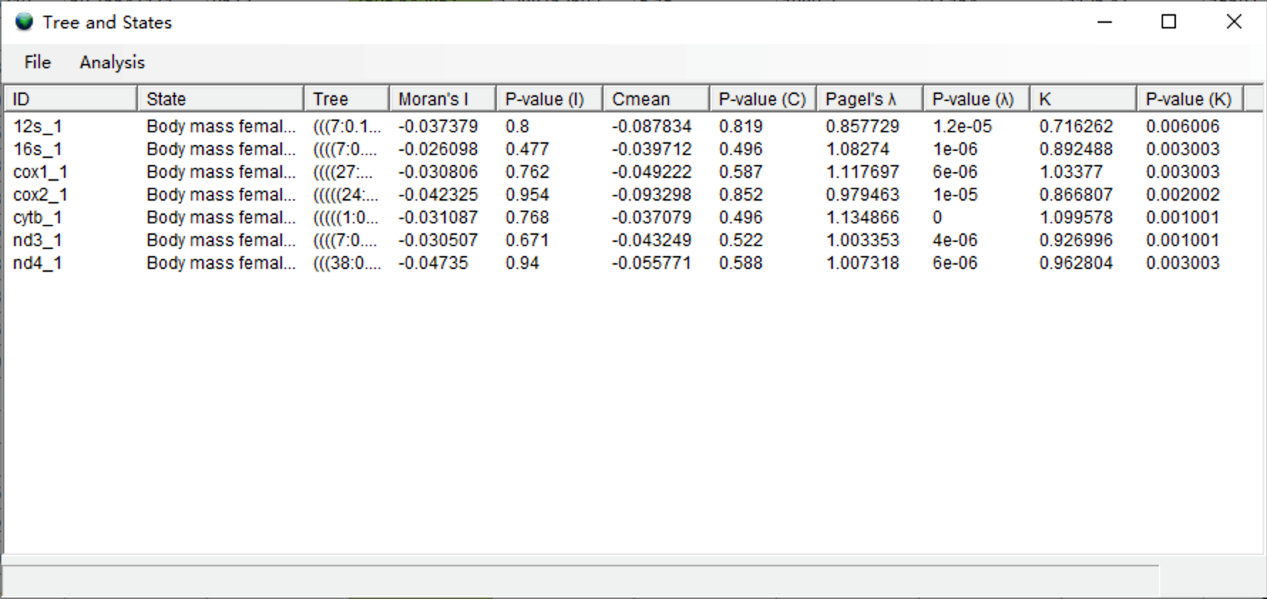
In the results, K values greater than 1 indicate strong phylogenetic signal that perfectly follows Brownian motion, and traits are phylogenetically conserved. The genes cox1 and cytb have the highest K values. This probably reflects a more similar evolutionary history of "Brain mass female mean" and these two genes.
NOTE: Because results using K and Pagel's λ were very similar, and K can also inform us about trait variation that is more similar than expected under Brownian motion, we focus on the results using K in this tutorial.
NOTE: Estimating phylogenetic signal of selected states on genes is not part of the original paper that analyzed these data. We simply wish to illustrate one of the new functionalities of RASP.
NOTE: The estimation of phylogenetic signal does not support the random trees function on the main RASP interface.
Estimate phylogenetic signal on states with consensus tree
Click [Analysis> Tree vs. States] to run the analysis. Users may see a command window (in Windows) with some additional information. Keep it open until it closes automatically. This analysis took one minute to run. When finished, you will see a window like the following:
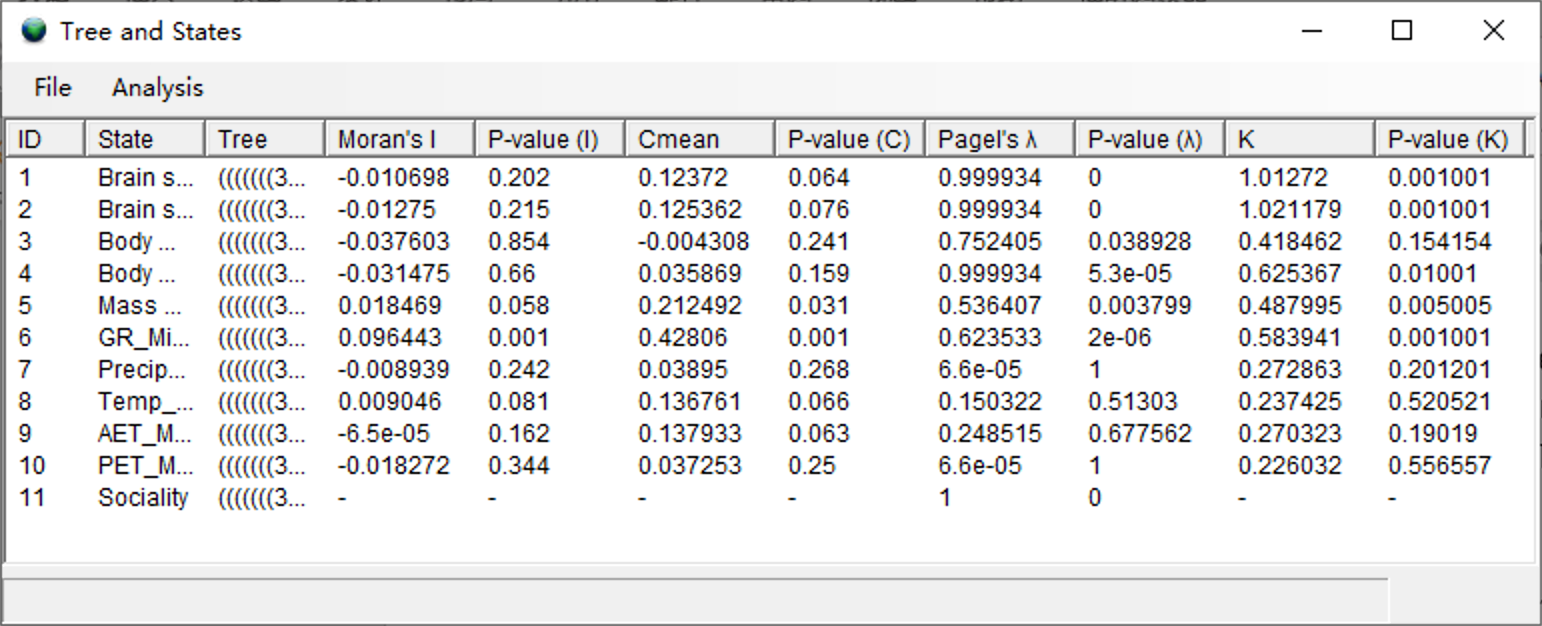
The high K and Pagel's λ values of brain sizes (ID 1 and 2) reflect the relatively invariable brain sizes within genera, and even within families, of primates (Kamilar and Cooper, 2013).
Open [File> Export> Full Table] to save the results.
References
Arnold, C., Matthews, L. J., & Nunn, C. L. (2010). The 10kTrees website: a new online resource for primate phylogeny. Evolutionary Anthropology: Issues, News, and Reviews, 19(3), 114-118.
Kamilar, J. M. & Cooper, N. (2013). Phylogenetic signal in primate behaviour, ecology and life history. Philos Trans R Soc Lond B Biol Sci, 368(1618), 20120341.
Shultz, S., Opie, C., & Atkinson, Q. D. (2011). Stepwise evolution of stable sociality in primates. Nature, 479(7372), 219-222.